Automatic segmentation of medical images is an important step to extract useful information that can help doctors make a diagnosis. For example, it can be used to segment retinal vessels so that we can represent their structure and measure their width which in turn can help diagnose retinal diseases.
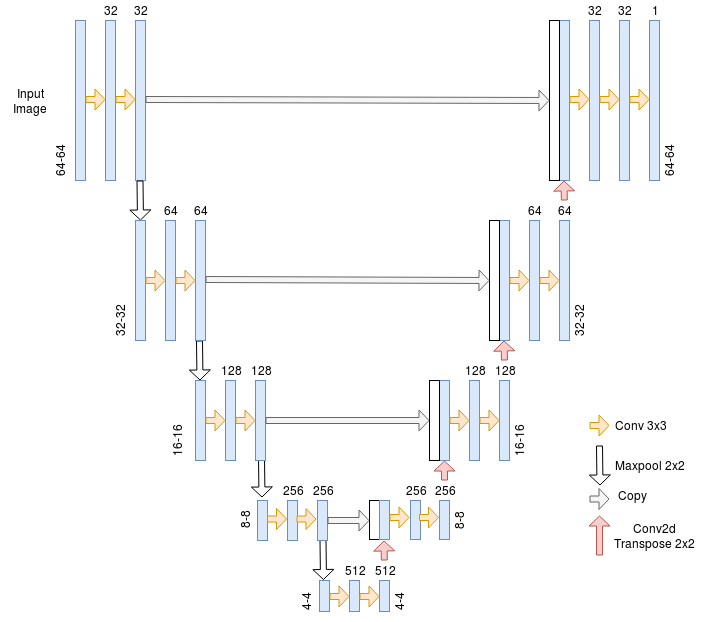
Here I implemented the U-net architecture to do blood vessel segmentation. It is an architecture that is widely used for semantic segmentation tasks especially in the medical domain.
Here I used DRIVE (Digital Retinal Images for Vessel Extraction) data set for all the experiments throughout the post. It is a data set of 40 retinal images ( 20 for training and 20 for testing ) where blood vessel were annotated at the pixel level ( see example above) to mark the presence (1) or absence (0) of a blood vessel at each pixel (i, j) of the image.
The U-net Architecture is an encoder-decoder with some skip connections between the encoder and the decoder. The major advantage of this architecture is its ability to take into account a wider context when making a prediction for a pixel. This is thanks to the large number of channels used in the up-sampling operation.
- Normalization : divided pixel intensities by 255 so they are in the 0–1 range.
- Cropping : The network expects each dimension of the input image to be divisible by 2⁴ because of the pooling operations so we take a random crop of 64*64 from each image.
- Data augmentation : Random flip (Horizontal or vertical or both), Random Shear, Random translation (Horizontal or vertical or both), Random Zoom. Performed during training only.
I trained two variations of the model :
- Pre-trained on ImageNet VGG encoder with data augmentation.
- Trained from scratch without data augmentation.